Biophysics 204B: Methods in Macromolecular Structure
Winter 2024 Syllabus
Course Title: Methods in Macromolecular Structure
Course Format: 6 hours of lecture/group work per week in class, substantial group work outside of class hours
Location and Date/Hours: Monday, Tuesday, Wednesday - 9AM-11AM in Genentech Hall 227
Prerequisites: All incoming first year BP and CCB graduate students are required to enroll in this course.
Grading: Letter grade
Textbook: None. Lab protocols and course materials will be available in class or online
Instructors: John Gross, Aashish Manglik, James Fraser, Klim Verba
TAs:
- Kevin.Delgado-Cunningham@ucsf.edu
- Tracy.Lou@ucsf.edu
- Henry.Scott@ucsf.edu
- Yisheng.Zheng@ucsf.edu
Lecturers/Facilitators:
Klim Verba, John Gross, Yifan Cheng, Aashish Manglik, Robert Stroud, Tom Goddard
Background:
Fluency in multiple biophysical methods is often critical for answering mechanistic questions. Traditionally, students are exposed to the fundamentals of multiple techniques through lectures that cover the theory prior to exposure, for some, in analysis or data collection during lab rotations. However, this structure means that only students that rotate in specific labs gain hands-on-exposure, which could limit adventurous experiments in future years. To train the next generation of biophysicists at UCSF, we have decided to alter this traditional structure by creating “Macromolecular Methods”, a class that places emphasis on playing with data. We have designed Macromolecular Methods to be a team-based class where students develop their own analysis of real data that, in non-pandemic years, they have collected. The goal of this course is to provide students sufficient exposure to major methods in biophysics to enable them to pursue these more fully in their independent research.
Course Description:
This is a team-based class where students work in small groups develop their own analysis of real data. Statistical aspects of rigor and reproducibility in structural biology will be emphasized throughout lectures, journal club presentations, and hands-on activities. The website for the 2017, 2018, 2019, 2020 editions are available online.
Course expectations: This course will likely be different from most other courses you have taken before. Our expectation is that you will maximize your learning by being an active participant - there is significant out of class work required to enable this. Many learning opportunities will arise if you are willing to take advantage of the hands on experience and from those taking time to show you interesting aspects of the various methods. Data analysis will not follow a linear path - our expectation is that you start to become confortable with unknowns and messy results. It is likely that you will encounter issues using the various scientific software required for the course - while we will help troubleshoot, navigating these challenges effectively is a core expectation that we want to see students build towrads during the course.
Ethics: This course is more than a training experience; data analysis is part of ongoing active research projects, the results of which will be published to the broader scientific community. The community must be able to understand our work, replicate it, and have confidence in its findings. We must therefore ensure the integrity of the information we disseminate. To do so, it is essential that students perform and document their experiments and analyses as faithfully as possible. Mistakes and oversights are normal and to be expected, but they must not be ignored, concealed, or disguised.
Respect: This course is built around an open research project performed in teams. Successful completion of the course objectives will require that students work together effectively, so please respect the time and effort of your classmates and instructors. Moreover, as part of the research process, we will consider and debate a variety of ideas and approaches; however, we must not allow our position on a particular idea or argument to compromise our respect for its author. We therefore expect course participants to give all instructors and students, regardless of academic or personal background, their complete professional respect; anything less will not be tolerated.
Accommodations for students with disabilities: The Graduate Division embraces all students, including students with documented disabilities. UCSF is committed to providing all students equal access to all of its programs, services, and activities. Student Disability Services (SDS) is the campus office that works with students who have disabilities to determine and coordinate reasonable accommodations. Students who have, or think they may have, a disability are invited to contact SDS (StudentDisability@ucsf.edu); or 415-476-6595) for a confidential discussion and to review the process for requesting accommodations in classroom and clinical settings. More information is available online at http://sds.ucsf.edu. Accommodations are never retroactive; therefore students are encouraged to register with Student Disability Services (http://sds.ucsf.edu/) as soon as they begin their programs. UCSF encourages students to engage in support seeking behavior via all of the resources available through Student Life, for consistent support and access to their programs.
Commitment to Diversity, Equity and Inclusion: The course instructors and teaching assistants value the contributions, ideas and perspective of all students. It is our intent that students from diverse backgrounds be well-served by this course, that students’ learning needs be addressed both in and out of class, and that the diversity that the students bring to this class be viewed as a resource, strength and benefit. It is our intent to present materials and activities that are respectful of diversity: gender identity, sexuality, disability, age, socioeconomic status, ethnicity, race, nationality, religion, and culture. However, we also acknowledge that many of the literature examples used in this course were authored in an environment that marginalized many groups. Integrating a diverse set of experiences is important for a more comprehensive understanding of science and we strive towards that goal. Although the instructors are committed to continuous improvement of our practices and our learning environment, we value input from students and your suggestions are encouraged and appreciated. Please let the course director or program leadership know ways to improve the effectiveness of the course for you personally, or for other students or student groups. (modeled after CCB and Brown University’s Diversity & Inclusion Syllabus Statements)
2023 schedule
TEAM ASSIGNMENTS
- Team name: A
- Gysasu Bajracharya
- Jules Brunello
- Hunter Carrell
- Yifei Chen
- Maple Chen
- Karson Chrispens
- Team name: B
- Zoey Dingman
- Kevin Alexander Estrada Alamo
- Haoyu Fan
- Amy Flis
- David Gomez Siu
- Katherine Hansen
- Team name: C
- William Ho
- Javarcia Ivory
- Jasmine Keyes
- Divya Kranthi
- Sean Myers
- Ever O’Donnell
- Sang Le
- Team name: D
- Angelica Lam
- Andreas Langen
- Ashley Lasko
- Mihn Le
- Kyrellos Ibrahim
- Luke Jaskowski
- Team name: E
- Zack Mawaldi
- Katie Holland
- Dru Myerscough
- Mingcheng Li
- Sruthi Raguveer
- Patrick Grimes
- Luke Villarma
Journal Club assignments
Jan 8-10 - Class intro
Monday January 8
- Course overview (AM)
- Introduction to instructors and TAs
- Structure of the class - timeline, topics covered, self-assessments
- Journal clubs
- Participation - teams and work-together recommendations/expectations
- Auditing
- Relationship to Macro mini-quals
- Final writeups
- High-level review of thermodynamics and kinetics (AM)
- Why study methods in biophysics? (AM)
- Few minute introduction to techniques covered
- Cryo-EM (KV)
- X-ray Crystallography (AM)
- NMR (JG)
- Out of class assignment (required!)
- Out of class suggested reading: Forces contributing to the conformational stability of proteins
Tuesday January 9
- An intuitive understanding of the Fourier Transform (AM)
- Fourier transform lecture (JG)
- Fourier transform in the different methods (JG/AM/KV)
- Resolution: start thinking about 3D objects like an X-ray or EM map, building intuition of more waves measured giving higher resolution
- Building up the MTZ (index = frequency and direction, amplitude/intensity, phase) and the concept of Nyquist frequency (why pixel size, changing values across pixels, and maximum resolution are related in EM)
- Understanding NMR signals as frequency domain information
- Software check the following:
- coot
- phenix
- ccp4
- chimerax
- pymol
- EMAN2
- NMRBox
- Out of class suggested material:
Wednesday January 10
Jan 16-24 - CryoEM - Lectures Yifan Cheng, Tutorials Klim Verba
Monday January 15
MLK DAY - HOLIDAY
Tuesday January 16
Wednesday January 17
Hands on practical day with David Bulkley on cryo-EM grid freezing
- 9:00-9:10AM: Journal club
Meet in Genentech Hall S101 at your team’s specified time.
- 9:10-9:40 AM: Teams 1&2
- 9:45-10:15 AM: Teams 3&4
- 10:20-10:50 AM: Team 5
Monday January 22
Hands on practical day with Klim and David on negative stain
- 40 minutes per team
- Upstairs is Genentech S416 with Klim Verba; Downstairs is Genentech S101 with David Bulkley
- 9:00-9:40: Team 1 with Daivd and Team 2 with Klim)
- 9:40-10:20: Team 3 with David and Team 4 with Klim
- 10:20-11:00: Team 5 with Klim
Tuesday January 23
Wednesday January 24
KV talk about resolution metrics/validation 9-10:30, catch up on processing 10:30-11.
Reading on rigor and reproducibility in EM:
Jan 29-Feb 7 X-ray Crystallography - Lectures Bob Stroud, Tutorials Aashish Manglik
Monday January 29th
9-1030: Lecture 1 from Bob Stroud
- What is the system?
- Examine diffraction data in adxv
Tuesday January 30th
Beamline trip/X-ray facility tour
- Beamline trip logistics
- If not going to beamline, please meet Violla Bassim in X-ray facility (Genentech S127) at 9AM
Wednesday January 31st
Beamline trip/X-ray facility tour
- Beamline trip logistics
- If not going to beamline, please meet Violla Bassim in X-ray facility (Genentech S127) at 9AM
Monday February 5
9-10:30: Lecture 2 from Bob Stroud
- Use xia2 to process diffraction data
- Understand various metrics for data reduction
- What do we have at the end? MTZ files!
- XRayView Download
Tuesday February 6
9-10:30: Lecture 3 from Bob Stroud
- File formats and contents
- MTZ file format
- PDB file as text
- Refinement as minimizing Fo-Fc residual
- Where do the phases come from?
- Molecular replacement
- Iterative phase improvement
- ASU vs. unit cell vs. monomer
- 2Fo-Fc map
- Fo-Fc map
- Iterative phase improvement affects density everywhere
- R-free
- B-factors
- The curse of high resolution!!!
- Model building
- Fixing stuff in real space
- Real space vs. reciprocal space refinment
- Prior knowledge (chemistry/physics) vs. Data in minimizing residual
- Water placement in Phenix or Coot
- Alternative conformations and partial occupancy
- conformational vs. compositional heterogeneity
- Team 1+2
- Team 3-5
- Alternative conformations of loops
- Occupancy refinement
- A/B conformations
- mutually exclusive
Wednesday February 7
9-10:30: Lecture 4 from Bob Stroud
Reading on rigor and reproducibility in Crystallography:
Feb 12-Feb 21 NMR - Lectures John Gross, Tutorials Amy Guo and Catherine Kuhn
Monday February 12
Tuesday February 13
- process 15N HSQC and 13C HSQC data with teams (Allie Born and John Gross)
Wednesday February 14
Monday February 19
Presidents Day - HOLIDAY
Tuesday February 20
Wednesday February 21
Lecture 5 from John Gross, Measuring ns-ps dynamics in proteins
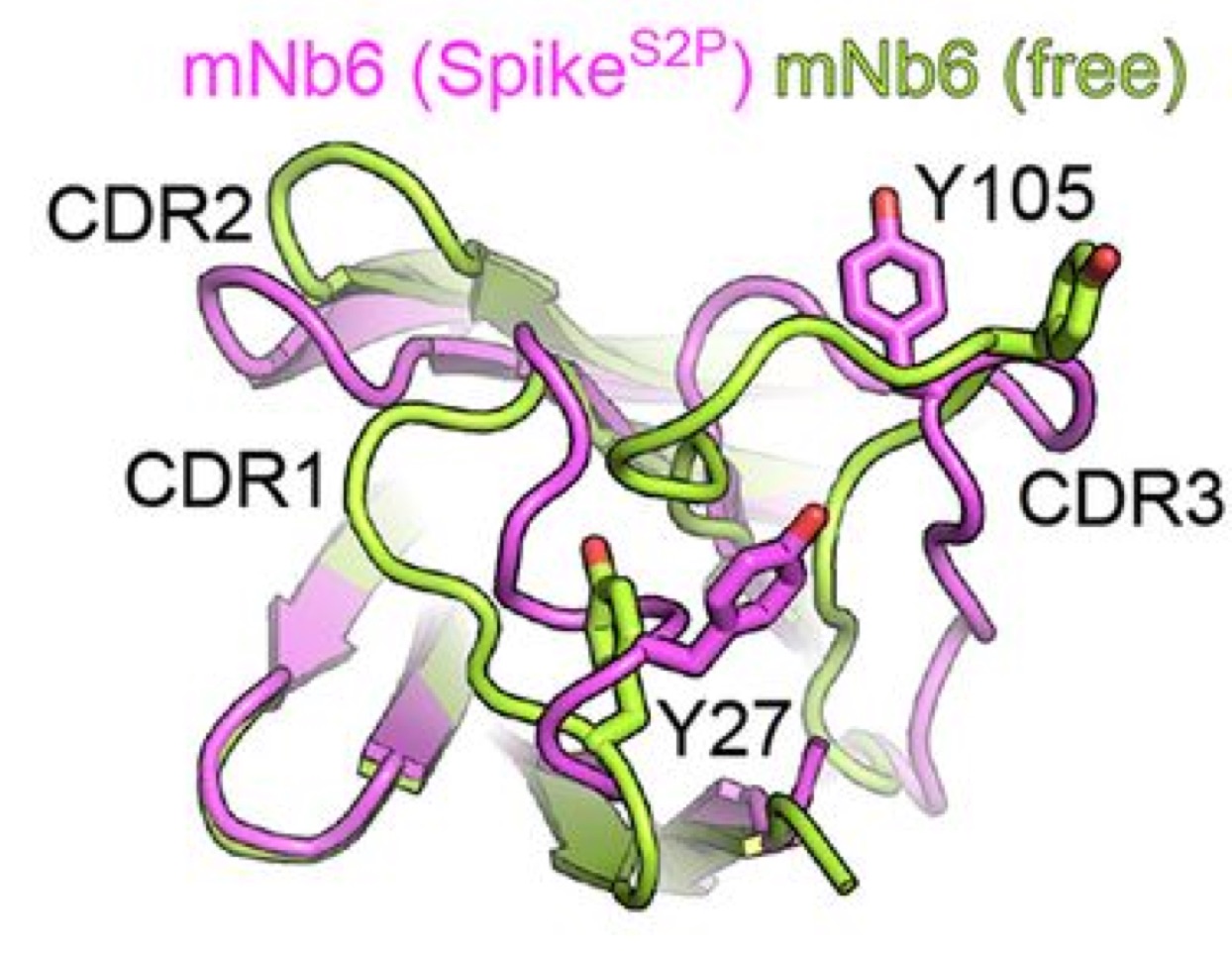
- process mNb6:RBD 15N and 13C HSQC spectra, overlay with mNb6. Cross-peak bookkeeping with teams (Allie Born and John Gross)
How to setup and process HSQCs on the 800 MHz spectrometer
Materials for TA Office Hours
Reading on rigor and reproducibility in NMR:
Final write up due: one per team - Feb 26
- EM: which of the 12 sequences/structure is your sample? Explain how you processed your data and identified which one it is? How can you quantify whether you are correct?
- Piezo1
- Spliceosome
- Apoferritin
- HSP104
- Cas9
- PEAK3/14-3-3 complex
- GroEL
- E. Coli Ribosome
- 20S Proteosome
- LKB1-StradA-Mo25 complex
- TRPV1
- Her2/Her3 complex
- Glutamine Synthetase
- SARS-CoV-2 Spike
- SARS CoV2 Nsp2
- BRAF:MEK:14-3-3 Complex
- X-ray: Introduce the problem you are trying to answer. Give a summary of data processing and refinement statistics (Table 1 type information). Provide results and interpretation of the ligand modeling excercise. How does the density in the binding site compare across your datasets? What roadblocks did you encounter, what did you try, how would you ideally model this? If you were successful in modeling the data, how did you quantify your data (presence/absence/occupancy of ligands)? If you weren’t successful, what is your interpretation? What conclusions can you draw about the forces driving ligand binding at the atomistic level? What would you do next?
- NMR: What is the NMR evidence that the nano bodies are undergoing conformational exchange in solution? Is the conformational change local or global? (Hint: where do the isoleucines in Nb6 and mNb6 map onto the structure?). Can you infer any differences between the dynamics of ancestral and mature Nb6 based on the NMR data? How does binding to Spike RBD change the dynamics? You may simply use the 13C HSQC on ILVMA labelled nanobodies to answer these questions though there is complementary information in the 15N HSQC spectra you may call on to support.
Supplemental material and tutorial videos