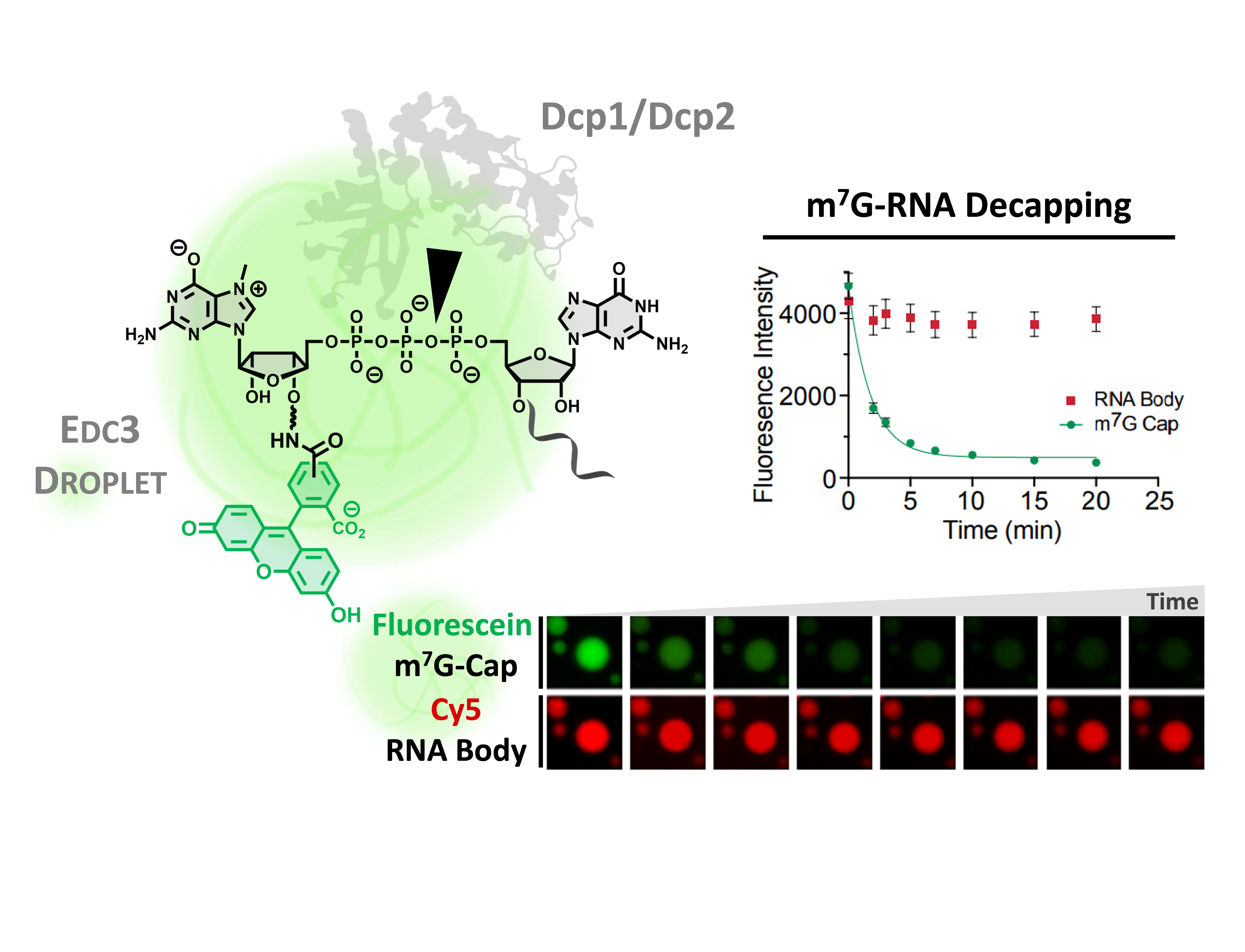
Approach for measuring mRNA decapping in biological condensates using dual fluorescent labeled RNA.
Biological condensates or membraneless organelles are conserved in all kingdoms of life. Condensate formation is driven by liquid-liquid phase separation. 5’-3’ decay factors localize to cytoplasmic mRNA processing bodies (P-bodies), which are conserved from yeast to humans. What is the impact of P-bodies on the function of 5’-3’ decay? The answer to this question is not clear, with some studies suggesting P-bodies are sites of mRNA storage whereas other reports indicate P-bodies are sites of mRNA decay. In collaboration with Drs Jacek Jemielity and Joanna Kowalska at the University of Warsaw we have developed approaches for measuring rates of enzyme catalyzed reactions in reconstituted condensates containing Dcp1/Dcp2 , substrate RNA and enhancers of decapping (EDCs). The long-term goal of this work is to take bottom-up approaches to investigate how composition and material properties affect decapping and subsequent steps of 5’-3’ decay in P-bodies.
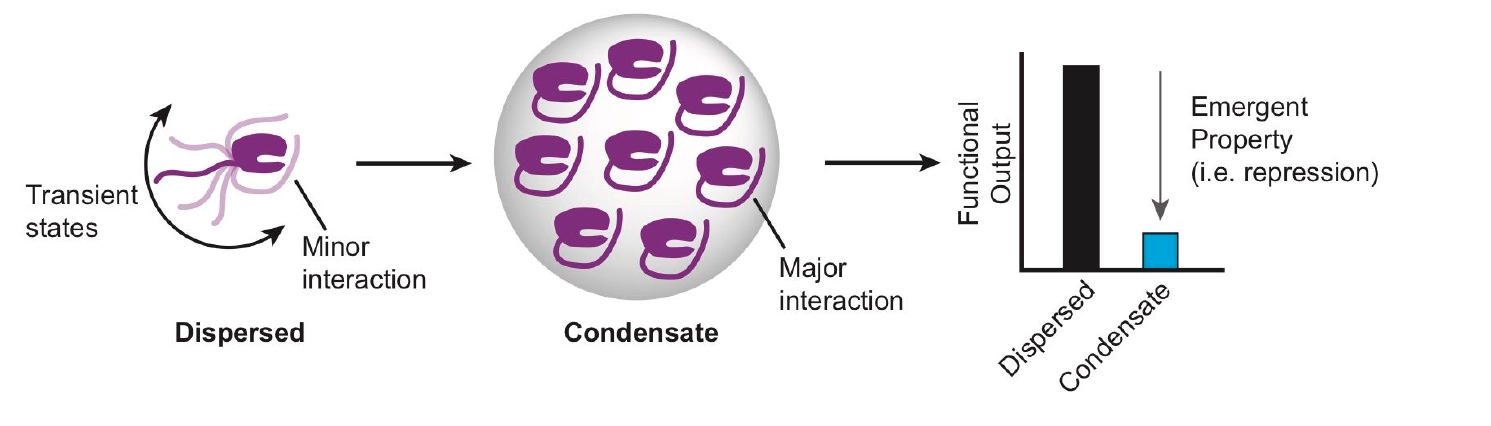
Model, liquid-liquid phase separation imparts emergent biological properties by conformational selection.
For example, we recently discovered that liquid-liquid phase separation reduces enzyme activity (100-fold), contrary to popular models positing that LLPS enhances enzyme activity by local concentration effects Tibble et al, Nature Chem Biol. Dynamic NMR provided evidence that the conformational landscape of the decapping complex in solution may be consolidated by interactions that promote liquid-liquid phase separation. A working model is that conformational landscapes of enzymes may be biased by liquid-liquid phase separation through conformational selection in the same way the crystallization can select for one or more conformations of a macromolecule present in solution. Understanding how emergent properties of condensates tune function of regulatory pathways is an important problem in cellular biochemistry, with implication for disease and drug discovery. Studying this problem requires an integrated approach including NMR spectroscopy.