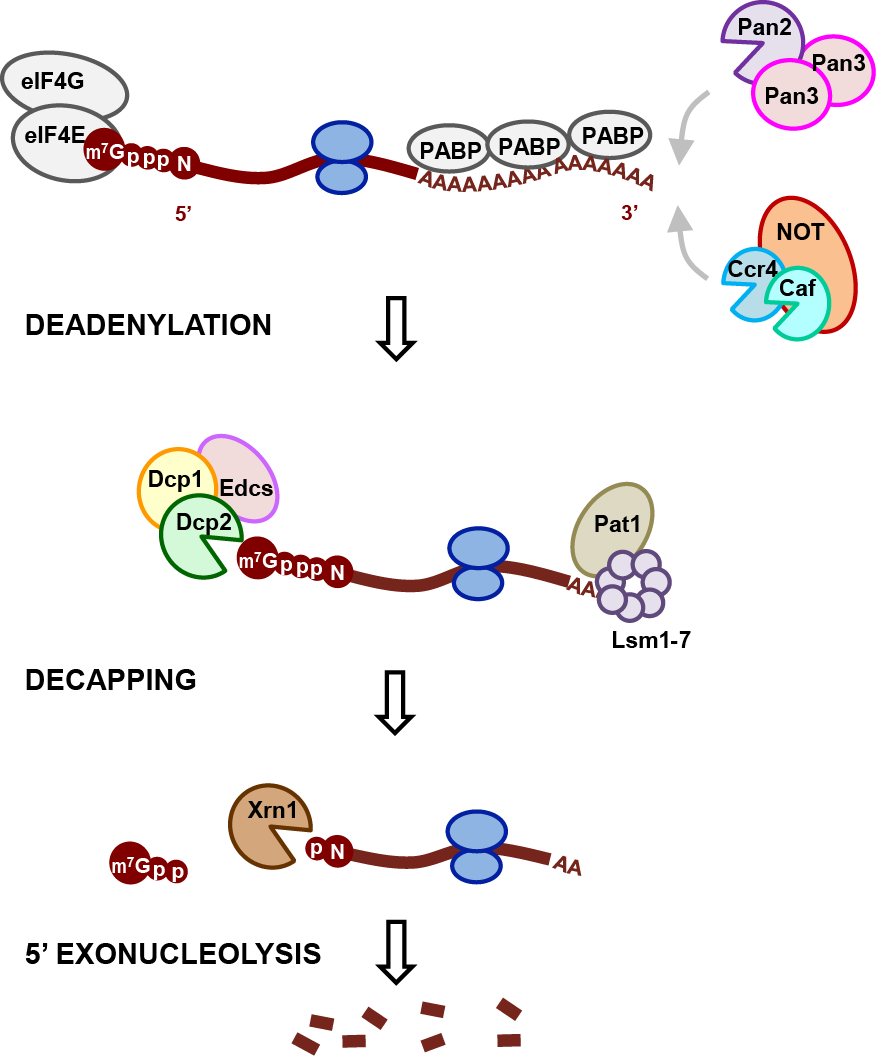
Major steps in the 5’-3’ RNA decay pathway.
The regulation of eukaryotic mRNA degradation plays a crucial role in many biological processes such as development, stress response, and gene expression. One major pathway used to control cellular mRNA stability is 5’-to-3’ mRNA decay, which requires removal of the protective 5’-terminal N7-methyl-guanosine (m7G) cap found on all eukaryotic mRNA. Hydrolysis of the m7G cap is catalyzed by the decapping enzyme Dcp2, in complex with the essential activator Dcp1 and decapping coactivators such as Edc1-4, Pat1/Lsm1-7, and Dhh1. Protein-protein interactions with coactivators modulate decapping activity and target the dynamic, multiprotein decapping complex to specific mRNA features and mRNA decay pathways. The decapping complex plays a key role in the control of RNA stability during deadenylation-dependent decay, nonsense-mediated decay, and microRNA-mediated decay. However, we lack a structure-level understanding of how decapping coactivators tune decapping activity on RNA and link decapping to cellular processes like 3’ deadenylation and translation by the ribosome.
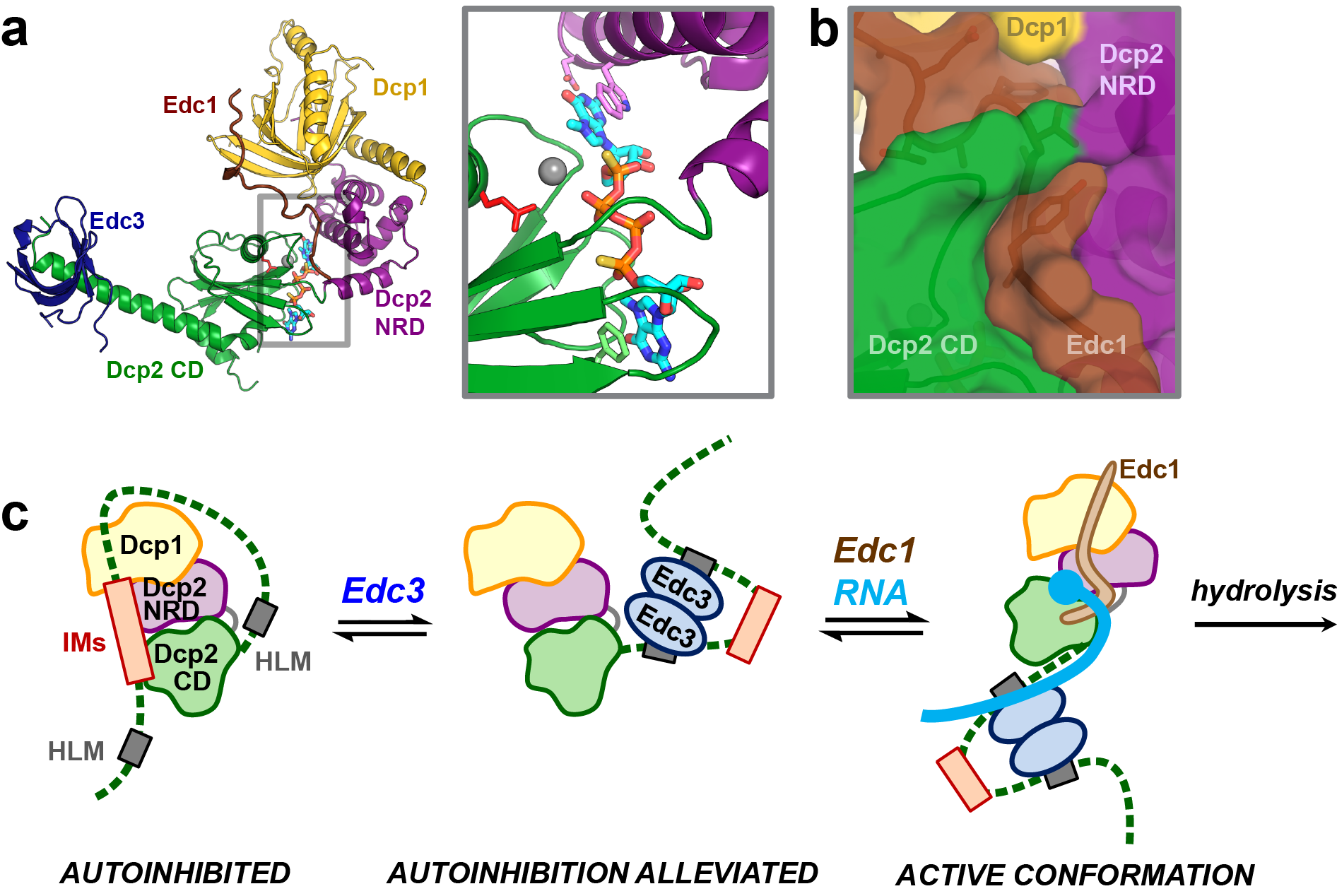
(a) Structure of the Edc1-Dcp1-Dcp2-Edc3 decapping complex in the active conformation with 5’ cap analog bound in the composite active site. (b) Coactivator Edc1 generates a 4-way protein-protein interaction to stabilize the active conformation and accelerate decapping. (c) Coactivators engage the disordered C-terminus of Dcp2 to alleviate autoinhibition and promote decapping.
Our lab combines a broad selection of experimental techniques that include NMR spectroscopy, X-ray crystallography, small-angle X-ray scattering, fluorescence anisotropy, in vitro decapping kinetics, and yeast genetics, in order to understand the links between the structure and conformation of the decapping complex with its cellular functions. Recent work in the Gross Lab has shown how Dcp2 uses a composite active site to bind and recognize the 5’ cap substrate and how Edc1-like coactivators accelerate catalysis by stabilizing the active conformation of the decapping complex (Mugridge et al Nat. Commun. 2018). We have also recently shown that the disordered C-terminal tail of yeast Dcp2 contains motifs that autoinhibit decapping activity and that decapping coactivators that bind within the C-terminal tail can alleviate this autoinhibition to turn on decapping (Paquette et al Nucleic Acids Res. 2018). We propose a model for regulation of mRNA decapping activity in which: (1) the Dcp1/Dcp2 decapping complex exists in an inactive, autoinhibited conformation in the absence of coactivators and substrate; (2) coactivators engage the decapping complex to alleviate autoinhbition and recruit the active decapping complex to target mRNA transcripts; and (3) coactivators promote conformational change into the active conformation of Dcp2 that uses a composite active site to catalyze cap cleavage. Building on this model, we are currently working to understand the structural and molecular mechanisms that link activation of 5’ end decapping to 3’ end deadenylation.